Introduction
Human leukocyte antigen (HLA) is closely involved in regulating the human immune system.
Despite the development of some methods for predicting classical HLA binders. Disappointingly, there are few methods or toolkits for predicting non-classical HLA binders due to a lack of experimental data.
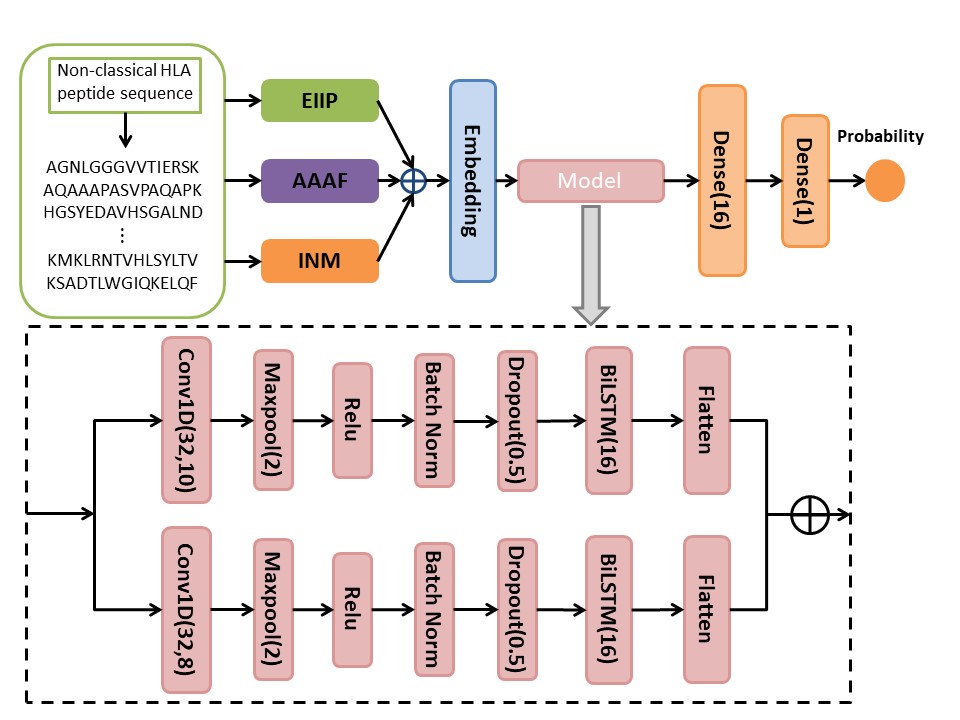
To advance research in the field of non-classical HLA binders, we have developed a deep learning-based tool called DeepHLApred, which fuses three features, Electron-ion interaction pseudo potential (EIIP),
Integer numerical mapping (INM), Accumulated amnio acid frequency (AAAF), and inputs them into a parallel convolutional neural network (CNN) and bi-directional long short term memory network (Bi-LSTM) architecture via an embedding layer.
The final prediction result is obtained through two fully connected layers, and has been proven to perform better and more balanced than the state-of-the-art methods,
regardless of whether the positive and negative samples are balanced or imbalanced. Furthermore, we have developed a webserver platform (www.biolscience.cn/DeepHLApred/) that is available for use by researchers.